Python BEAM

Original logo produced by Oscar Adams CC BY-SA 4.0
BEAM (Broad EPOCH Analysis Modules) is a collection of independent yet complementary open-source tools for analysing EPOCH simulations in Python, designed to be modular, allowing researchers to adopt only the components they require without being constrained by a rigid framework. In line with the FAIR principles — Findable, Accessible, Interoperable, and Reusable — each package is openly published with clear documentation and versioning (Findable), distributed via public repositories (Accessible), designed to follow common standards for data structures and interfaces (Interoperable), and includes licensing and metadata to support long-term use and adaptation (Reusable).
- sdf-xarray: Processing and plotting of SDF files and converting them to xarray.
- epydeck: Input deck reader and writer.
- epyscan: Create campaigns over a given parameter space using various sampling methods.
Originally developed by Joel Adams and the PlasmaFAIR Team at the York Plasma Institute under the EPSRC Grant EP/V051822/1.
Installation
All of the packages are available on PyPI and can be installed using pip:
pip install sdf-xarray epydeck epyscan
Each package can be used independently or combined, depending on your needs.
Citation
If any of the BEAM packages contribute to a project that leads to publication, please acknowledge this by citing the module in question. This can be done by clicking the “Cite this repository” button located near the top right of their respective GitHub pages.
Contribution
We welcome contributions to the BEAM ecosystem! Whether it’s reporting issues, suggesting features, or submitting pull requests, your input helps improve these tools for the community. Please follow the contribution guidelines in each repository and feel free to reach out via GitHub discussions or issues.
sdf-xarray: analysing EPOCH output
sdf-xarray is a lightweight wrapper that reads EPOCH’s SDF files into xarray.Dataset objects. This provides:
- Lazy loading and memory-efficient handling of large datasets
- Easy slicing and plotting using
matplotlib,xarray.plot, orhvplot - Automatic normalisation of grid and variable names
- Integration with Jupyter notebooks and Dask for parallel analysis
The key features for this module are highlighted below, for in-depth documentation please visit https://sdf-xarray.readthedocs.io
Single file loading
import xarray as xr
ds = xr.open_dataset("0010.sdf")
ds["Electric_Field_Ex"]
# <xarray.DataArray 'Electric_Field_Ex' (X_x_px_deltaf_electron_beam: 16)> Size: 128B
# [16 values with dtype=float64]
# Coordinates:
# * X_x_px_deltaf_electron_beam (X_x_px_deltaf_electron_beam) float64 128B 1...
# Attributes:
# units: V/m
# full_name: "Electric Field/Ex"
Multi file loading
To open a whole simulation at once, pass preprocess=sdf_xarray.SDFPreprocess()
to xarray.open_mfdataset:
import xarray as xr
from sdf_xarray import SDFPreprocess
ds = xr.open_mfdataset("*.sdf", preprocess=SDFPreprocess())
print(ds)
# Dimensions:
# time: 301, X_Grid_mid: 128, ...
# Coordinates: (9) ...
# Data variables: (18) ...
# Indexes: (9) ...
# Attributes: (22) ...
SDFPreprocess checks that all the files are from the same simulation, as
ensures that there’s a time dimension so the files are correctly concatenated.
If your simulation has multiple output blocks so that not all variables are
output at every time step, then those variables will have NaN values at the
corresponding time points.
After loading a series of datasets, we can select a simulation file by calling the .isel() function and passing the parameter time=0, where 0 can be any number between 0 and the total number of simulation files.
We can also use the .sel() function if we know the exact simulation time we want to select. There must be a corresponding dataset with this time for it to work correctly.
print(f"There are a total of {ds["time"].size} time steps. (This is the same as the number of SDF files in the folder)")
# There are a total of 41 time steps. (This is the same as the number of SDF files in the folder)
print(f"The time steps are: {ds["time"].values}")
# The time steps are: [2.60596949e-17 5.00346143e-15 1.00069229e-14, ..., 2.00034218e-13]
sim_time = ds['time'].isel(time=20).values
print(f"The time at the 20th simulation step is {sim_time:.2e} s")
# The time at the 20th simulation step is 1.00e-13 s
ds["Electric_Field_Ex"].isel(time=20)
# OR
# ds["Electric_Field_Ex"].sel(time=sim_time)
Plotting
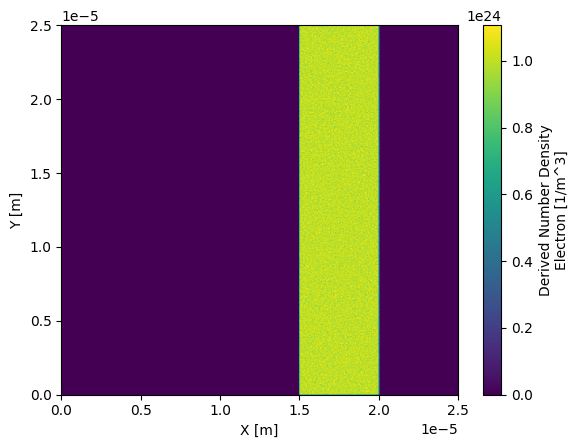
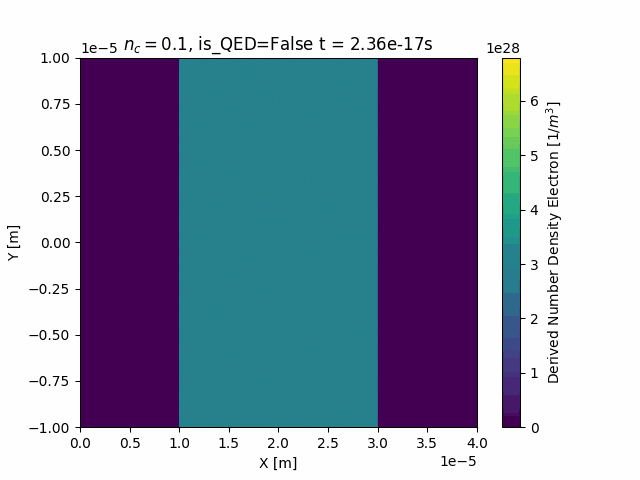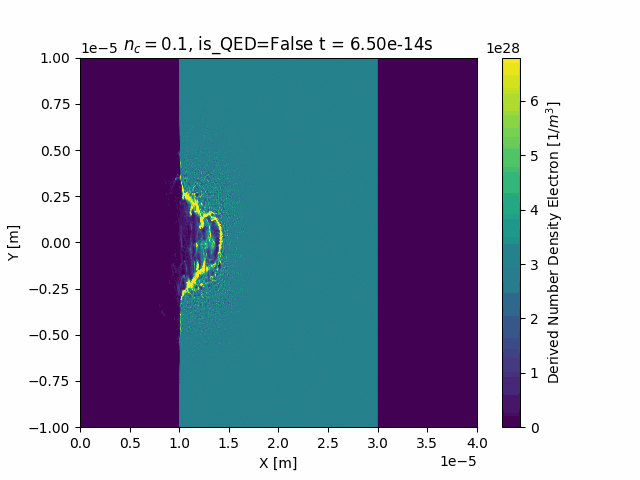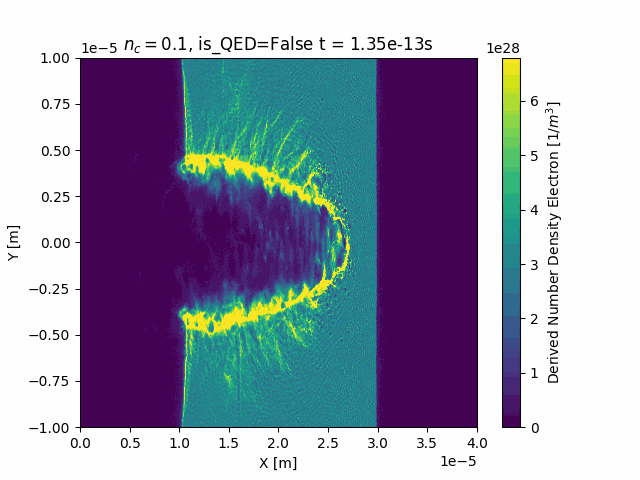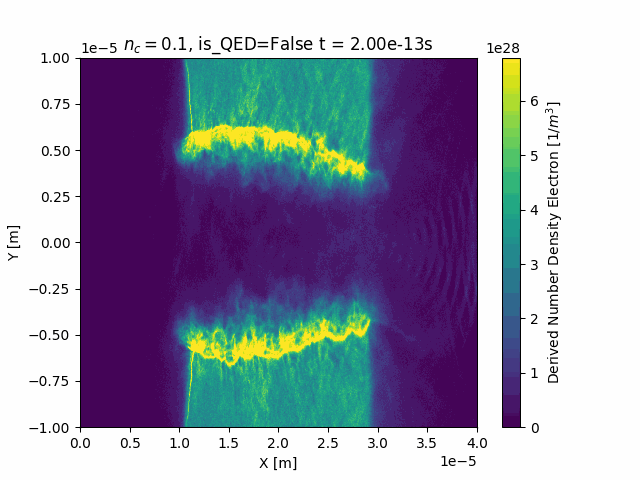
Since this package converts SDF files to xarray, we can leverage the plotting features that are included in xarray. For example we can load in a SDF file and plot the number density of the electrons:
import xarray as xr
ds = xr.open_dataset("0010.sdf")
# NOTE: EPOCH saves the x and y axes in SDF files in the inverse order of what is expected by xarray, so we must specify which axis is which; otherwise, our plot will be inverted.
ds["Derived_Number_Density_Electron"].plot(x="X_Grid_mid", y="Y_Grid_mid")
Animating
This package also contains custom functionality to generate time-resolved animations by loading in all the SDF files in a given simulation run:
import xarray as xr
from sdf_xarray import SDFPreprocess
ds = xr.open_mfdataset("*.sdf", preprocess=SDFPreprocess())
ani = ds["Derived_Number_Density_Electron"].epoch.animate()
ani.save("Derived_Number_Density_Electron.gif", fps=5)
epydeck: Writing input deck files with Python
Writing a large number of EPOCH input files by hand can be tedious and error-prone. epydeck (short for EPOCH Python deck) allows you to create and manipulate input decks in Python:
- Build decks using standard Python data structures
- Load, modify, and save EPOCH-style
.deckfiles - Designed to preserve comments and formatting where possible
The interface follows the standard Python
json module:
epydeck.loadto read from afileobjectepydeck.loadsto read from an existing stringepydeck.dumpto write to afileobjectepydeck.dumpsto write to a string
The key features for this module are highlighted below, for in-depth documentation please visit https://github.com/epochpic/epydeck
Example
import epydeck
# Read from a file with `epydeck.load`
with open(filename) as f:
deck = epydeck.load(f)
print(deck.keys())
# dict_keys(['control', 'boundaries', 'constant', 'species', 'laser', 'output_global', 'output', 'dist_fn'])
# Modify the deck as a usual python dict:
deck["species"]["proton"]["charge"] = 2.0
# Write to file
with open(filename, "w") as f:
epydeck.dump(deck, f)
print(epydeck.dumps(deck))
# ...
# begin:species
# name = proton
# charge = 2.0
# mass = 1836.2
# fraction = 0.5
# number_density = if((r gt ri) and (r lt ro), den_cone, 0.0)
# number_density = if((x gt xi) and (x lt xo) and (r lt ri), den_cone, number_density(proton))
# number_density = if(x gt xo, 0.0, number_density(proton))
# end:species
# ...
epyscan: Campaign generation and parameter sampling
epyscan (short for EPOCH Python scan) generates
EPOCH campaigns over a parameter space using different sampling methods. It supports the following features:
- Defining scans over one or more input variables
- Support for grid and Latin hypercube sampling methods
Parameter space to be sampled is described by a dict where keys
should be in the form of block_name:parameter, and values should
be dicts with the following keys:
"min": minimum value of the parameter"max": maximum value of the parameter"log": (optional)bool, ifTruethen grid is done in log space for this parameter
The key features for this module are highlighted below, for in-depth documentation please visit https://github.com/epochpic/epyscan
Example
import pathlib
import epyscan
import epydeck
# Define the parameter space to be sampled. Here, we are varying the intensity
# and density
parameters = {
# Intensity varies logarithmically between 1.0e22 and 1.0e24
"constant:intens": {"min": 1.0e22, "max": 1.0e24, "log": True},
# Density varies logarithmically between 1.0e20 and 1.0e24
"constant:nel": {"min": 1.0e20, "max": 1e24, "log": True},
}
# Load a deck file to use as a template for the simulations
with open("template_deck_filename") as f:
deck = epydeck.load(f)
# Create a grid scan object that will generate 4 different sets of parameters
# within the specified ranges
grid_scan = epyscan.GridScan(parameters, n_samples=4)
# Define the root directory where the simulation folders will be saved.
# This directory will be created if it does not already exist
run_root = pathlib.Path("example_campaign")
# Initialize a campaign object with the template deck and the root directory.
# This will manage the creation of simulation cases
campaign = epyscan.Campaign(deck, run_root)
# Generate the folders and deck files for each set of parameters in the
# grid scan
paths = [campaign.setup_case(sample) for sample in grid_scan]
# Save the paths of the generated simulation folders to a file
with open("paths.txt", "w") as f:
[f.write(f"{path}\n") for path in paths]
# Example content of paths.txt
# example_campaign/run_0_1000000/run_0_10000/run_0_100/run_0
# example_campaign/run_0_1000000/run_0_10000/run_0_100/run_1
# example_campaign/run_0_1000000/run_0_10000/run_0_100/run_2
# ...